Projects
We are a Computational Biology lab that focuses on methods’ development and inference in evolutinary biology and specifically in phylogenetics. We strive to understand how different organisms evolve over long time periods by making meaningful inferences about thier evolutinary histories using state-of-the-art approaches of machine learning.
Computational phylogenetics
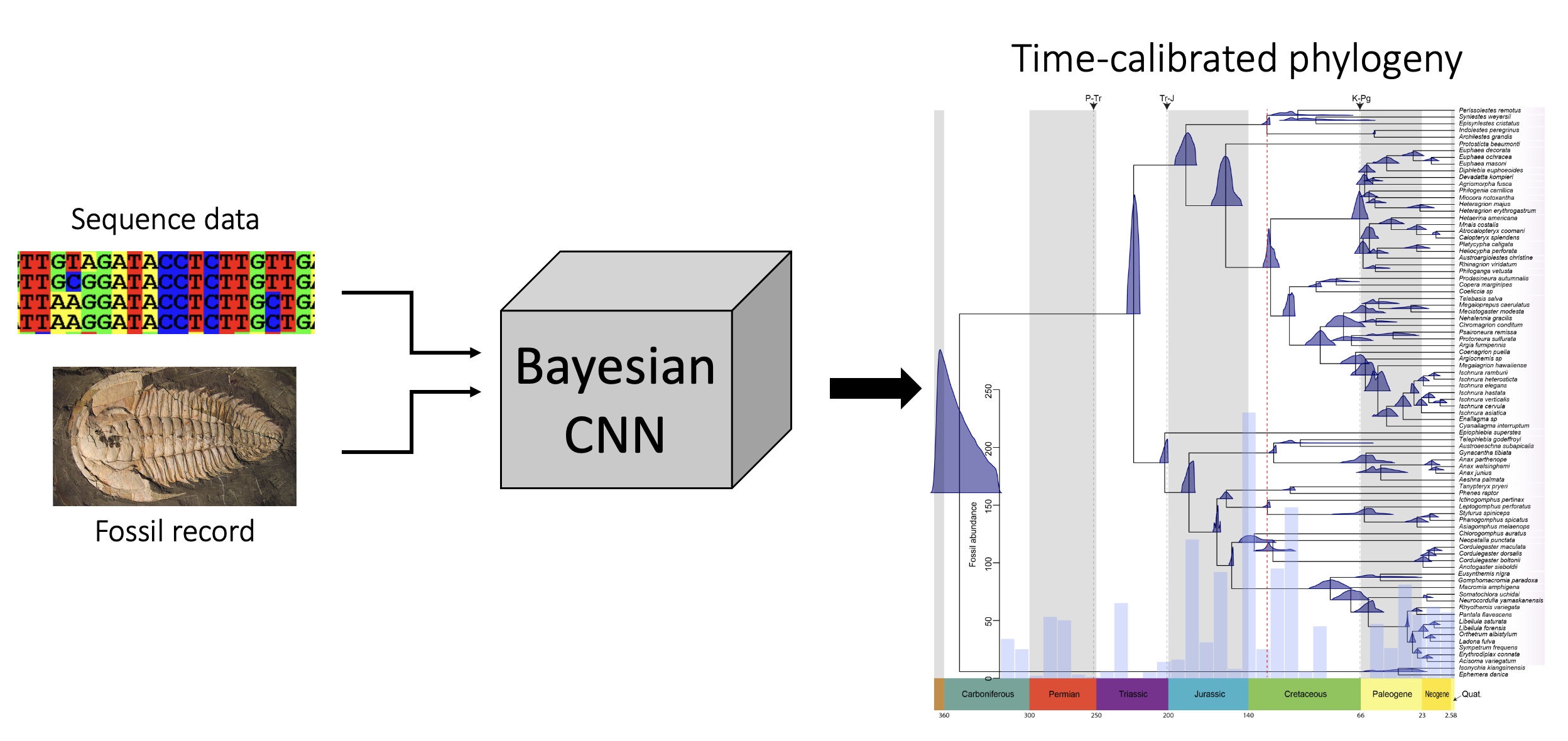
1. Developing new tree-dating approaches using Bayesian Neural Networks.
This project is concentrated on the development of new deep learning approaches for dating phylogenetic trees, i.e. estimating branch lengths in absolute time units that correspond to Earth’s geological timeline. This calss of methods will be capable of using external information such as fossil calibration points, mutation rates and/or biogeographic data to time-calibrate phylogenies.
2. Inference of phylogenetic networks using deep neural learning
In recent years there a large body of evidence has been amassed that demonstrates that multiple parts of the Tree of Life did not evolve according to a strictly bifurcating phylogeny. Instead, many organisms experience reticulate network-like evolution that is caused by an exchange of inter-specific genetic information. Thus, this project will include development of a convolutional neural network (CNN) that is capable of distinguishing different phylogenetic reticulation scenarios caused by introgression. More specifically, this will involve inferring the correct topology of phylogenetic networks from multi-locus sequence data.
Empirical phylogenomics
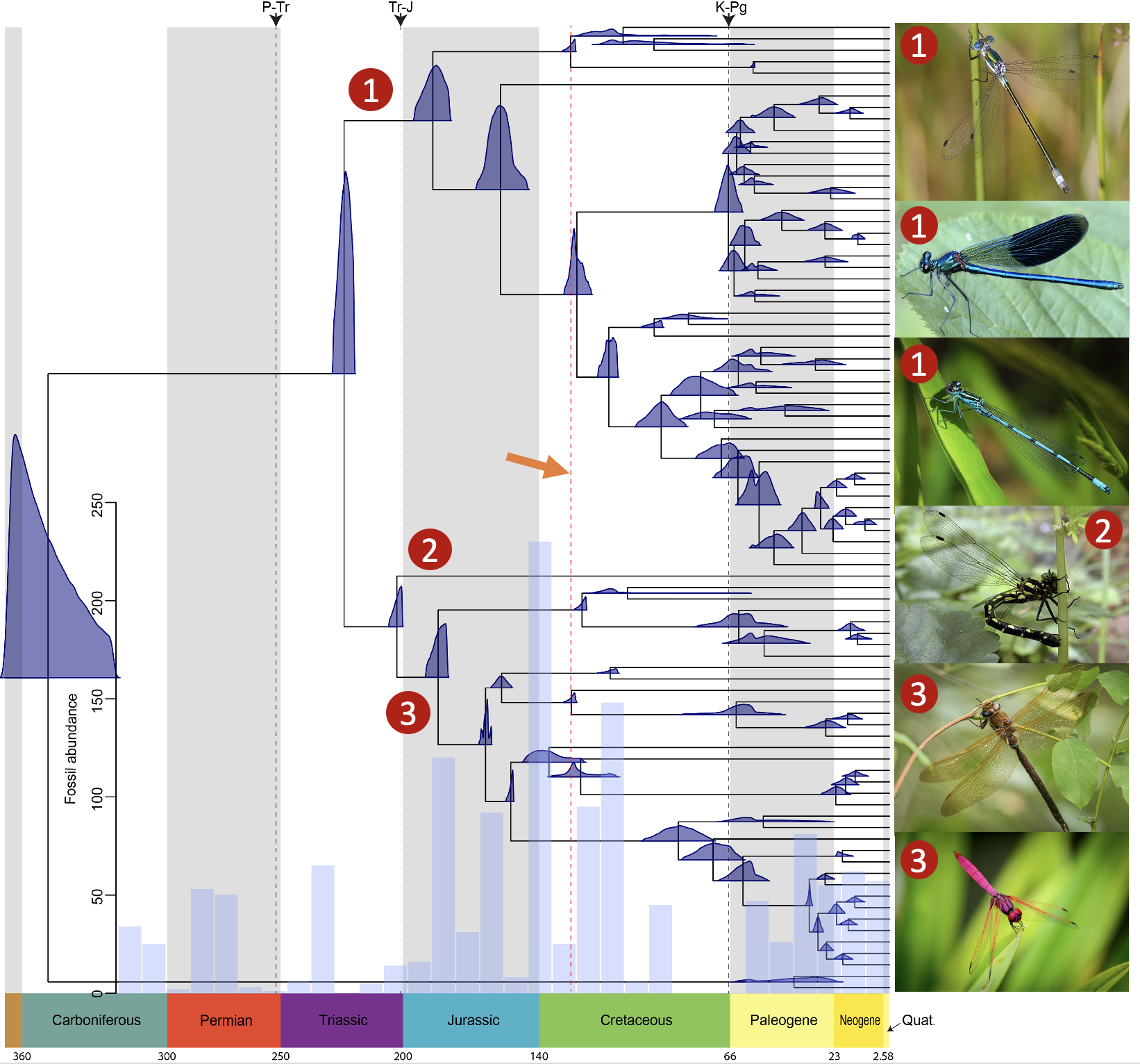
1. Phylogenomics of odoanta
Odonata (dragonflies and damselflies) represent a unique empirical system containing >6,300 extant species with one of the largest and complete fossil records in the animal kingdom. These data represent a remarkable opportunity to tackle such fundamental questions as the evolution of animal flight, mass migration, color vision, past and ongoing extinction events and their effect on paleo- and contemporary biodiversity, and the occurrence of ancient introgression. This project will aim to generate several representative highly contiguous genome assemblies across all three odonate suborders and reconcile them with fossil information and existing heterogeneous genetic data. During this project our lab will be actively collaborating with Mark Blaxter in the Tree of Life Programme at Sanger Institute.
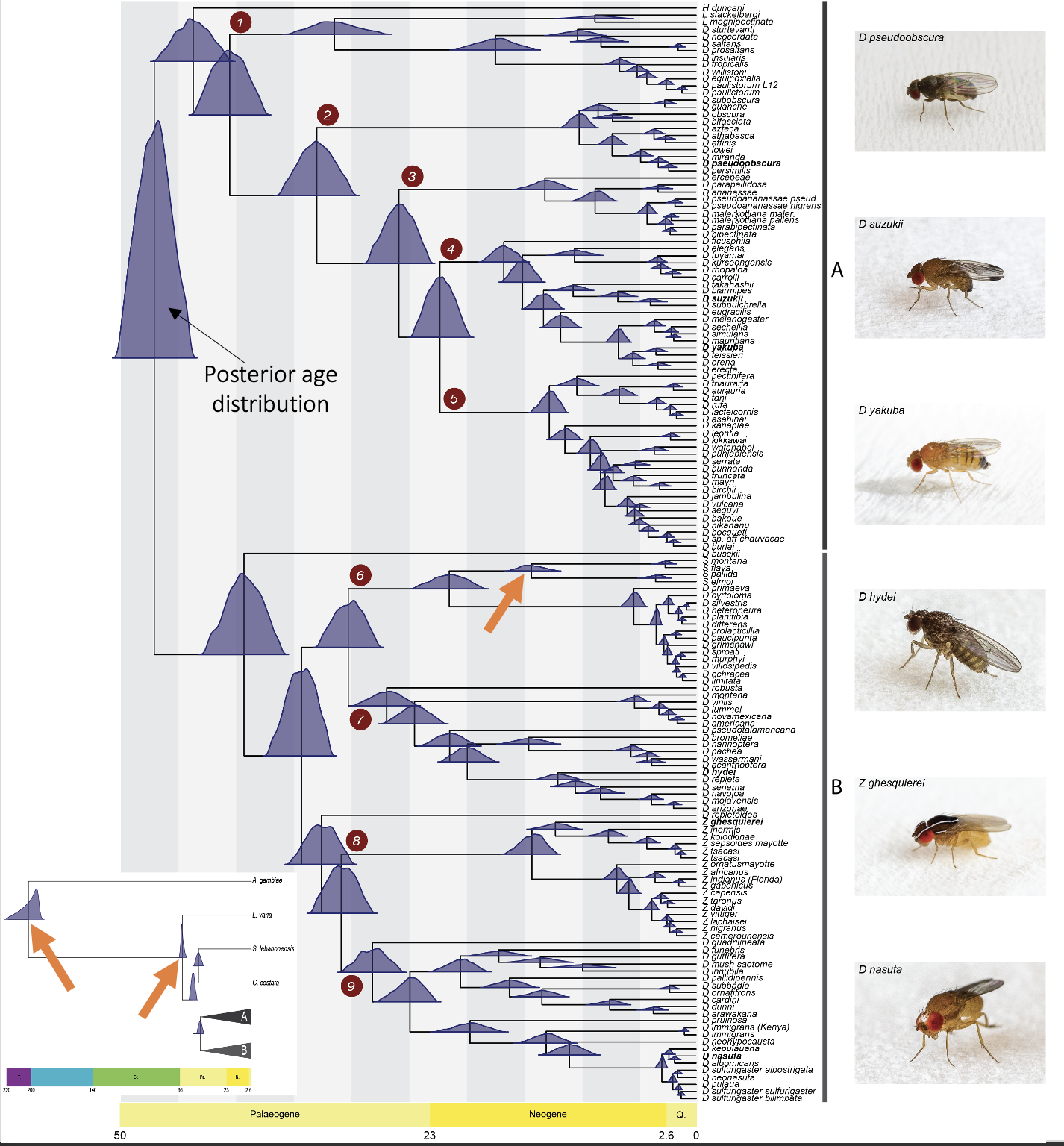
2. Drosophilidae Tree of Life
Our recent work has shown that, contrary to prior beliefs that post-speciation gene flow is relatively rare in Drosophila, introgression is in fact pervasive across the evolutionary history of the genus. However, the extent of introgression is unknown beyond the genus level. In close collaboration with Bernard Kim at Stanford Univeristy, this project will focus on phylogenetic estimates of family-level Drosophilidae evolutionary history and patterns of post-speciation gene flow using phylogenomic methods.